Perspectives on ENCODE
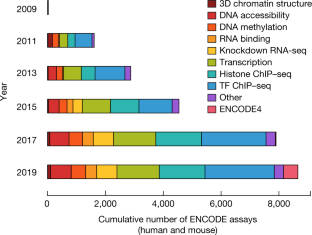
The Encylopedia of DNA Elements (ENCODE) Project launched in 2003 with the long-term goal of developing a comprehensive map of functional elements in the human genome. These included genes, biochemical regions associated with gene regulation (for example, transcription factor binding sites, open chromatin, and histone marks) and transcript isoforms. The marks serve as sites for candidate cis-regulatory elements (cCREs) that may serve functional roles in regulating gene expression 1 . The project has been extended to model organisms, particularly the mouse. In the third phase of ENCODE, nearly a million and more than 300,000 cCRE annotations have been generated for human and mouse, respectively, and these have provided a valuable resource for the scientific community.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
196,21 € per year
only 3,85 € per issue
Buy this article
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others

Expanded encyclopaedias of DNA elements in the human and mouse genomes
Article Open access 29 July 2020
Gapped-kmer sequence modeling robustly identifies regulatory vocabularies and distal enhancers conserved between evolutionarily distant mammals
Article Open access 31 July 2024

The RNA Atlas expands the catalog of human non-coding RNAs
Article 17 June 2021
Change history
References
- Kellis, M. et al. Defining functional DNA elements in the human genome. Proc. Natl Acad. Sci. USA111, 6131–6138 (2014). ArticleADSCASGoogle Scholar
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science306, 636–640 (2004). ArticleADSGoogle Scholar
- Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature478, 476–482 (2011). ArticleCASGoogle Scholar
- Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature420, 520–562 (2002). ArticleADSCASGoogle Scholar
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 (2011). ArticleGoogle Scholar
- Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature447, 799–816 (2007). The results of the pilot phase of ENCODE included extensive functional assays across a selected one per cent of the human genome with experiments conducted on a variety of cell lines and largely with array-based technology. ArticleADSCASGoogle Scholar
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). The results of the second phase of ENCODE were based mostly on a large number of genome-wide assays that leveraged high-throughput sequencing technologies and were done across two ‘tier one’ cell lines with large-scale assays across several hundred cell and tissue types. ArticleADSGoogle Scholar
- The ENCODE Project Consortium et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Naturehttps://doi.org/10.1038/s41586-020-2493-4 (2020).
- Partridge, E. C. et al. Occupancy maps of 208 chromatin-associated proteins in one human cell type. Naturehttps://doi.org/10.1038/s41586-020-2023-4 (2020).
- Meuleman, W. Index and biological spectrum of human DNase I hypersensitive sites. Naturehttps://doi.org/10.1038/s41586-020-XXXX-X (2020).
- Vierstra, J. et al. Global reference mapping of human transcription factor footprints. Naturehttps://doi.org/10.1038/s41586-020-2528-x (2020).
- Breschi, A. et al. A limited set of transcriptional programs define major cell types. Preprint at https://doi.org/10.1101/857169 (2020).
- Grubert, F. et al. Landscape of cohesin-mediated chromatin loops in the human genome. Naturehttps://doi.org/10.1038/s41586-020-2151-x (2020).
- Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA binding proteins. Naturehttps://doi.org/10.1038/s41586-020-2077-3 (2020).
- Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). ArticleCASGoogle Scholar
- Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods5, 621–628 (2008). ArticleCASGoogle Scholar
- Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein–DNA interactions. Science316, 1497–1502 (2007). ArticleADSCASGoogle Scholar
- Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods4, 651–657 (2007). ArticleCASGoogle Scholar
- Iyer, V. R. et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature409, 533–538 (2001). ArticleADSCASGoogle Scholar
- Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science290, 2306–2309 (2000). ArticleADSCASGoogle Scholar
- Landt, S. G. et al. ChIP–seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). A consortium-wide effort to standardize performance, quality control and outputs of ChIP–seq experiments, including validation of antibodies, to facilitate experimental reproducibllity and data utility. ArticleCASGoogle Scholar
- Sundararaman, B. et al. Resources for the comprehensive discovery of functional RNA elements. Mol. Cell61, 903–913 (2016). ArticleCASGoogle Scholar
- Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science337, 1190–1195 (2012). ArticleADSCASGoogle Scholar
- Schaub, M. A., Boyle, A. P., Kundaje, A., Batzoglou, S. & Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012). ArticleCASGoogle Scholar
- Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature515, 355–364 (2014). Results of a large-scale effort of the mouse ENCODE consortium, presenting regulatory and transcript maps of the mouse. ArticleADSCASGoogle Scholar
- Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science330, 1775–1787 (2010). ArticleADSCASGoogle Scholar
- The modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science330, 1787–1797 (2010). ArticleGoogle Scholar
- Kudron, M. M. et al. The ModERN Resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics208, 937–949 (2018). ArticleCASGoogle Scholar
- Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Naturehttps://doi.org/10.1038/s41586-020-2093-3 (2020).
- He, P. A. The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Naturehttps://doi.org/10.1038/s41586-020-XXXX-X (2020).
- He, Y. et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Naturehttps://doi.org/10.1038/s41586-020-2119-x (2020).
- Cheng, Y. et al. Principles of regulatory information conservation between mouse and human. Nature515, 371–375 (2014). ArticleADSCASGoogle Scholar
- Stefflova, K. et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell154, 530–540 (2013). ArticleCASGoogle Scholar
- Keilwagen, J., Posch, S. & Grau, J. Accurate prediction of cell type-specific transcription factor binding. Genome Biol. 20, 9 (2019). ArticleGoogle Scholar
- Tang, F., Lao, K. & Surani, M. A. Development and applications of single-cell transcriptome analysis. Nat. Methods8 (Suppl), S6–S11 (2011). ArticleCASGoogle Scholar
- Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature523, 486–490 (2015). ArticleADSCASGoogle Scholar
- Hu, B. C.; HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature574, 187–192 (2019). ArticleADSGoogle Scholar
- Regev, A. et al. The human cell atlas. eLife6, e27041 (2017). ArticleGoogle Scholar
- Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics13, 278–289 (2015). ArticleGoogle Scholar
- Arnold, C. D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science339, 1074–1077 (2013). ArticleADSCASGoogle Scholar
- Klein, J. C., Chen, W., Gasperini, M. & Shendure, J. Identifying novel enhancer elements with CRISPR-based screens. ACS Chem. Biol. 13, 326–332 (2018). ArticleCASGoogle Scholar
- Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). ArticleCASGoogle Scholar
- Paudyal, A. et al. The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC Dev. Biol. 10, 87 (2010). ArticleGoogle Scholar
Acknowledgements
We thank S. Moore, E. Cahill, M. Kellis and J. Li for their assistance, and B. Wold for helpful comments. This work was supported by grants from the NIH: U01HG007019, U01HG007033, U01HG007036, U01HG007037, U41HG006992, U41HG006993, U41HG006994, U41HG006995, U41HG006996, U41HG006997, U41HG006998, U41HG006999, U41HG007000, U41HG007001, U41HG007002, U41HG007003, U41HG007234, U54HG006991, U54HG006997, U54HG006998, U54HG007004, U54HG007005, U54HG007010 and UM1HG009442.
Author information
Authors and Affiliations
- Department of Genetics, School of Medicine, Stanford University, Palo Alto, CA, USA Nicholas J. Addleman, Jessika Adrian, Ulugbek K. Baymuradov, Anand Bhaskar, Nathan Boley, Lulu Cao, Hassan Chaib, Esther T. Chan, Songjie Chen, J. Michael Cherry, Timothy R. Dreszer, Idan Gabdank, Fabian Grubert, Nastaran Heidari, Jason A. Hilton, Benjamin C. Hitz, Lixia Jiang, Maya Kasowski, Trupti Kawli, Daniel Sunwook Kim, Anshul Kundaje, Jin Wook Lee, Yang I. Li, Yining Li, Shin Lin, Tejaswini Mishra, Anil M. Narasimha, Aditi K. Narayanan, Kathrina C. Onate, Jonathan K. Pritchard, Anil Raj, Lucia Ramirez, Denis N. Salins, Eilon Sharon, Minyi Shi, Teri Slifer, Cricket A. Sloan, Michael P. Snyder, Damek V. Spacek, Rohith Srivas, J. Seth Strattan, Forrest Y. Tanaka, Oana Ursu, Nathaniel K. Watson, Xinqiong Yang, Jie Zhai, Michael P. Snyder, Benjamin C. Hitz & J. Michael Cherry
- Cardiovascular Institute, Stanford School of Medicine, Stanford, CA, USA Michael P. Snyder & Michael P. Snyder
- Functional Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA Cassidy Danyko, Carrie A. Davis, Alexander Dobin, Thomas R. Gingeras, Alexandra Scavelli, Lei Hoon See, Chris Zaleski & Thomas R. Gingeras
- University of Massachusetts Medical School, Program in Bioinformatics and Integrative Biology, Worcester, MA, USA Gregory R. Andrews, Tyler Borrman, Jason A. Brooks, Hao Chen, Shaimae I. Elhajjajy, Kaili Fan, Kevin Fortier, Yu Fu, Mingshi Gao, Jack Huey, Eugenio Mattei, Jill E. Moore, Nishigandha N. Phalke, Henry E. Pratt, Michael J. Purcaro, Thomas M. Reimonn, Shuo Shan, Junko Tsuji, Arjan G. van der Velde, Zhiping Weng, Taylor Young, Xiao-Ou Zhang, Jill E. Moore & Zhiping Weng
- Department of Thoracic Surgery, Clinical Translational Research Center, Shanghai Pulmonary Hospital, The School of Life Sciences and Technology, Tongji University, Shanghai, China Lingling Cai, Shaliu Fu, Shuyu Hou, Xiangrui Li, Ying Li, Yan Liu, Aiping Lu, Qin Wang, Zhijie Wei, Zhiping Weng, Tianxiong Yu, Peng Zhang & Zhiping Weng
- Bioinformatics Program, Boston University, Boston, MA, USA Hao Chen, Arjan G. van der Velde, Zhiping Weng & Zhiping Weng
- Yale University, New Haven, CT, USA Declan Clarke, Timur Galeev, Mark B. Gerstein, Mengting Gu, Gamze Gursoy, Arif Harmanci, Sushant Kumar, Donghoon Lee, Jason Liu, Shaoke Lou, Fabio C. P. Navarro, Baikang Pei, Joel Rozowsky, Anurag Sethi, Cristina Sisu, Jinrui Xu, Koon-Kiu Yan, Jing Zhang & Mark B. Gerstein
- Ludwig Institute for Cancer Research, University of California, San Diego, La Jolla, CA, USA Sora Chee, David U. Gorkin, Hui Huang, Samantha Kuan, Ah Young Lee, Bin Li, Sebastian Preissl, Bing Ren, Yin Shen, Yanxiao Zhang, Yuan Zhao & Bing Ren
- Center for Epigenomics, University of California, San Diego, La Jolla, CA, USA David U. Gorkin, Bing Ren & Bing Ren
- Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA, USA Belinda M. Giardine, Ross C. Hardison, Cheryl A. Keller, Maria Long & Ross C. Hardison
- Altius Institute for Biomedical Sciences, Seattle, WA, USA Reyes Acosta, Daniel Bates, Michael Buckley, Andres Castillo, Daniel R. Chee, Morgan Diegel, Douglass Dunn, Alister P. W. Funnell, Grigorios Georgolopoulos, Jessica Halow, Eric Haugen, Sean Ibarrientos, Mineo Iwata, Audra Johnson, Rajinder Kaul, Tanya Kutyavin, John Lazar, Kristen Lee, Wouter Meuleman, Vivek Nandakumar, Patrick Navas, Jemma Nelson, Shane Neph, Fidencio Jun Neri, Andrew Nishida, Ericka Otterman, Alex Reynolds, Eric Rynes, Richard Sandstrom, Anthony Shafer, Kyle Siebenthall, John A. Stamatoyannopoulos, Sandra Stehling-Sun, Benjamin Van Biber, Jeff Vierstra, Shinny Vong, Hao Wang, Robert E. Welikson, Yongqi Yan & John A. Stamatoyannopoulos
- Department of Genome Sciences, University of Washington, Seattle, WA, USA William S. Noble, John A. Stamatoyannopoulos, Galip Gürkan Yardımcı & John A. Stamatoyannopoulos
- Department of Medicine, University of Washington, Seattle, WA, USA Rajinder Kaul, John A. Stamatoyannopoulos & John A. Stamatoyannopoulos
- Department of Genetics and Genome Sciences, Institute for Systems Genomics, UConn Health, Farmington, CT, USA Michael O. Duff, Brenton R. Graveley, Sara Olson, Xintao Wei, Lijun Zhan & Brenton R. Graveley
- National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA Omar Al Jammal, Eileen Cahill, Julie Coursen, Elise A. Feingold, Daniel A. Gilchrist, Samuel H. Moore, Preetha Nandi, Hannah Naughton, Briana Nuñez, Michael Pagan, Michael J. Pazin, Jeffery A. Schloss, Yekaterina Vaydylevich, Simona Volpi, Xiao-Qiao Zhou, Elise A. Feingold, Michael J. Pazin, Michael Pagan & Daniel A. Gilchrist
- Broad Institute and Department of Pathology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA Bradley E. Bernstein & Bradley E. Bernstein
- Biological Sciences, University of Alabama in Huntsville, Huntsville, AL, USA Surya B. Chhetri, Candice J. Coppola, Eric M. Mendenhall & Eric M. Mendenhall
- HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA Laurel A. Brandsmeier, Surya B. Chhetri, Candice J. Coppola, Rachel C. Evans, Shawn Levy, Mark Mackiewicz, Megan McEown, Sarah K. Meadows, Eric M. Mendenhall, Christopher L. Messer, Dianna E. Moore, Richard M. Myers, Amy R. Nesmith, J. Scott Newberry, Kimberly M. Newberry, Rosy Nguyen, E. Christopher Partridge, Florencia Pauli-Behn, Brian Roberts, Collin White, Eric M. Mendenhall & Richard M. Myers
- European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Cambridge, UK Bronwen Aken, If H. A. Barnes, Daniel Barrell, Ruth Bennett, Andrew Berry, Alexandra Bignell, Claire Davidson, Sarah Donaldson, Paul Flicek, Adam Frankish, Carlos Garcia Giron, Jose M. Gonzalez, Matthew Hardy, Toby Hunt, Osagie Izuogu, Michael Kay, Jane Loveland, Fergal J. Martin, Jonathan M. Mudge, Emily H. Perry, Marie-Marthe Suner, Daniel R. Zerbino, Daniel R. Zerbino, Adam Frankish & Paul Flicek
- The Broad Institute of Harvard and MIT, Cambridge, MA, USA Kristin G. Ardlie, Samantha Beik, Charles B. Epstein, Nina Farrell, Alon Goren, Meital Hatan, David Hendrickson, Shanna Hsu, Robbyn Issner, Irwin Jungreis, Manolis Kellis, Oren Ram, Joseph Raymond, Noam Shoresh, Danielle Tenen, Mia C. Uziel, Christopher M. Vockley & David Wine
- MGH, Boston, MA, USA Yotam Drier, Shawn Gillespie, Michael Mannstadt, Dylan Rausch, Miguel Rivera & Russell Ryan
- Dana-Farber Cancer Institute, Boston, MA, USA Mariateresa Fulciniti, Birgit Knoechel & Nikhil Munshi
- Harvard Medical School, Boston, MA, USA Qian Qin & Jonathan Scheiman
- Boston Children’s Hospital, Boston, MA, USA Birgit Knoechel
- Harvard University, Cambridge, MA, USA David Kelley
- Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, USA Carles Boix, Jose Davila-Velderrain, Matthew D. Edwards, David K. Gifford, Yuchun Guo, Tatsunori B. Hashimoto, Lei Hou, Tommi Jaakkola, Irwin Jungreis, Manolis Kellis, Pouya Kheradpour, Yue Li, Dianbo Liu, Yaping Liu, Eva Maria Novoa, Charles W. O’Donnell, Yongjin Park, Tahin Syed, Maxim Wolf, Haoyang Zeng & Zhizhuo Zhang
- Max Planck Institute for Molecular Genetics, Department of Genome Regulation, Berlin, Germany Alex Meissner
- University of Colorado Boulder, Boulder, CO, USA John Rinn
- Bioinformatics and Genomics Program, Centre for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology and Universitat Pompeu Fabra, Barcelona, Spain Beatrice Borsari, Alessandra Breschi, Sarah Djebali, Roderic Guigó, Rory Johnson, Julien Lagarde, Dmitri D. Pervouchine, Barbara Uszczynska-Ratajczak & Anna Vlasova
- IRSD, Université de Toulouse, INSERM, INRA, ENVT, UPS, U1220, CHU Purpan, CS60039, Toulouse, France Sarah Djebali
- Skolkovo Institute for Science and Technology, Moscow, Russia Dmitri D. Pervouchine
- Functional Genomics, Cold Spring Harbor Laboratory, Woodbury, NY, USA Jorg Drenkow
- Department of Clinical Research, University of Bern, Bern, Switzerland Rory Johnson
- International Institute of Molecular and Cell Biology, Warsaw, Poland Barbara Uszczynska-Ratajczak
- Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA Cassandra Bazile, Christopher B. Burge, Daniel Dominguez, Abigail Hochman, Nicole J. Lambert, Tsultrim Palden & Amanda Su
- Department of Cellular and Molecular Medicine, Institute for Genomic Medicine, Stem Cell Program, Sanford Consortium for Regenerative Medicine, University of California, San Diego, La Jolla, CA, USA Steven M. Blue, Keri Elkins, Chelsea Anne Gelboin-Burkhart, Thai B. Nguyen, Gabriel A. Pratt, Ines Rabano, Shashank Sathe, Rebecca Stanton, Balaji Sundararaman, Eric L. Van Nostrand, Ruth Wang, Brian A. Yee & Gene W. Yeo
- Département de Biochimie et Médecine Moléculaire, Université de Montréal, Montréal, Quebec, Canada Louis Philip Benoit Bouvrette, Neal A. L. Cody, Eric Lécuyer & Xiaofeng Wang
- Division of Experimental Medicine, McGill University, Quebec, Canada Louis Philip Benoit Bouvrette, Neal A. L. Cody, Eric Lécuyer & Xiaofeng Wang
- Institut de Recherches Cliniques de Montréal (IRCM), Montréal, Quebec, Canada Louis Philip Benoit Bouvrette, Neal A. L. Cody, Eric Lécuyer & Xiaofeng Wang
- Program in Computational and Systems Biology, Massachusetts Institute of Technology, Cambridge, MA, USA Peter Freese
- Department of Cellular and Molecular Medicine, Institute of Genomic Medicine, University of California, San Diego, San Diego, CA, USA Jia-Yu Chen, Xiang-Dong Fu, Hairi Li & Rui Xiao
- Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA Daniel Savic
- Division of Biology, California Institute of Technology, Pasadena, CA, USA Henry Amrhein, Igor Antoshechkin, Gilberto DeSalvo, Katherine Fisher-Aylor, Peng He, Anthony Kirilusha, Georgi K. Marinov, Kenneth McCue, Diane E. Trout, Sean A. Upchurch, Jost Vielmetter, Brian A. Williams & Barbara Wold
- National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA Stacie M. Anderson, David M. Bodine & Elisabeth F. Heuston
- Department of Biostatistics and Bioinformatics, Duke University, Durham, NC, USA Timothy E. Reddy
- Center for Genomic and Computational Biology, Duke University, Durham, NC, USA Anthony M. D’Ippolito & Timothy E. Reddy
- Biological Sciences, University of California, Irvine, Irvine, CA, USA Nicole El-Ali, Camden Jansen, Ali Mortazavi, Rabi Murad, Ricardo N. Ramirez & Weihua Zeng
- Salk Institute for Biological Studies, La Jolla, CA, USA Rosa G. Castanon, Huaming Chen, Joseph R. Ecker, Manoj Hariharan, Yupeng He, Graham McVicker & Joseph R. Nery
- Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA Veena Afzal, Jennifer A. Akiyama, Iros Barozzi, Diane E. Dickel, Yoko Fukuda-Yuzawa, Tyler H. Garvin, Anne Harrington, Momoe Kato, Elizabeth Lee, Brandon J. Mannion, Catherine S. Novak, Len A. Pennacchio, Quan Pham, Ingrid Plajzer-Frick, Valentina Snetkova & Axel Visel
- Department of Chemistry and Biochemistry, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA, USA Bo Ding, Nan Li, Vu Ngo, Tung Nguyen, Mengchi Wang, Tao Wang, Wei Wang, John W. Whitaker, Andre Wildberg, Kai Zhang & Yun Zhu
- Institute for Human Genetics, Department of Neurology, University of California, San Francisco, San Francisco, CA, USA Yin Shen
- Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Chicago, IL, USA Feng Yue
- Penn State Health Milton S. Hershey Medical Center, Hershey, PA, USA Tingting Liu, Yanli Wang, Jie Xu, Hongbo Yang, Feng Yue, Bo Zhang & Lijun Zhang
- US Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA Len A. Pennacchio & Axel Visel
- School of Natural Sciences, University of California, Merced, Merced, CA, USA Axel Visel
- Comparative Biochemistry Program, University of California, Berkeley, CA, USA Len A. Pennacchio
- Howard Hughes Medical Institute, The Salk Institute for Biological Studies, La Jolla, CA, USA Joseph R. Ecker
- Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI, USA Alan P. Boyle
- Department of Human Genetics, University of Michigan, Ann Arbor, MI, USA Alan P. Boyle
- Department of Molecular and Cellular Physiology, School of Medicine, Stanford University, Palo Alto, CA, USA Catharine Eastman
- Terrence Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario, Canada Andrew Emili, Jack F. Greenblatt, Hongbo Guo, Timothy R. Hughes, Ernest Radovani & Guoqing Zhong
- Department of Genetics, School of Medicine, Yale University, New Haven, CT, USA Peng Gong, Jin Lian, Xinghua Pan, Quan Shen, Sherman M. Weissman & Jialing Zhang
- Department of Biochemistry and Molecular Medicine, Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA Peggy J. Farnham, Yu Guo, Suhn K. Rhie & Heather N. Witt
- Department of Radiation Oncology, School of Medicine, Stanford University, Palo Alto, CA, USA Nastaran Heidari
- Department of Human Genetics, Institute for Genomics and Systems Biology, The University of Chicago, Chicago, IL, USA Cory Holgren, Nader Jameel, Madhura Kadaba, Mary Kasparian, Lijia Ma, Matthew G. Milton, Jennifer Moran, Dave Steffan, Matt Szynkarek, Dave Toffey & Alec Victorsen
- Division of General Surgery, Section of Transplant Surgery, School of Medicine, Washington University, St. Louis, MO, USA Yiing Lin
- Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, China Xinghua Pan
- Guangdong Provincial Key Laboratory of Single Cell Technology and Application, Guangzhou, China Xinghua Pan
- Department of Cell Biology & Physiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA Doug H. Phanstiel
- Thurston Arthritis Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA Doug H. Phanstiel
- Department of Molecular Genetics, University of Toronto, Toronto, Ontario, Canada Jack F. Greenblatt, Rozita Razavi & Frank W. Schmitges
- School of Medicine, Jiangsu University, Zhenjiang, China Quan Shen
- Department of Molecular Genetics, Donnelly Centre, University of Toronto, Toronto, Ontario, Canada Timothy R. Hughes
- Tempus Labs, Chicago, IL, USA Kevin P. White
- Department of Medicine, University of Washington, Seattle, WA, USA George Stamatoyannopoulos
- Fred Hutchinson Cancer Research Center, Seattle, WA, USA M. A. Bender, Rachel Byron & Mark T. Groudine
- HHMI and Program in Systems Biology, University of Massachusetts Medical School, Albert Sherman Center, Worcester, MA, USA Job Dekker, Bryan R. Lajoie, Hakan Ozadam & Ye Zhan
- University of Massachusetts Amherst, Amherst, MA, USA Yu Fu
- Institute for Infocomm Research, Singapore, Singapore Chuan Sheng Foo
- Simon Fraser University, Burnaby, British Columbia, Canada Maxwell W. Libbrecht
- Center for Functional Cancer Epigenetics, Dana-Farber Cancer Institute, Boston, MA, USA Peng Jiang, X. Shirley Liu, Clifford Meyer & Chongzhi Zang
- Department of Data Sciences, Dana-Farber Cancer Institute and Harvard T.H. Chan School of Public Health, Boston, MA, USA Peng Jiang, X. Shirley Liu, Clifford Meyer & Chongzhi Zang
- Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA Chongzhi Zang
- Molecular Pathology Unit & Cancer Center, Boston, MA, USA Qian Qin
- Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL, USA Mingxiang Teng
- Institute for Stem Cell Biology and Regenerative Medicine, Stanford University, Stanford, CA, USA Amira A. Barkal
- Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA Budhaditya Banerjee, Maria Gutierrez-Arcelus, Nisha Rajagopal, Soumya Raychaudhuri, Richard I. Sherwood, Sharanya Srinivasan & Harm-Jan Westra
- Department of Statistics, Medical Sciences Center, Madison, WI, USA Sunduz Keles, Jurijs Nazarovs & Ye Zheng
- Department of Biostatistics and Medical Informatics, Madison, WI, USA Colin N. Dewey, Sunduz Keles, Peng Liu & Rene Welch
- Department of Mathematical Sciences, University of Texas at Dallas, Richardson, TX, USA Sunyoung Shin
- Department of Statistics, University of Nebraska-Lincoln, Lincoln, NE, USA Qi Zhang
- Department of Biomedical Informatics, The Ohio State University, Columbus, OH, USA Dongjun Chung
- Department of Cell and Regenerative Biology, UW-Madison Blood Research Program, Carbone Cancer Center, University of Wisconsin School of Medicine and Public Health, University of Wisconsin, Madison, WI, USA Emery H. Bresnick
- Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA James E. Hayes & Robert J. Klein
- Wellcome Sanger Institute, Cambridge, UK Federico Abascal, Matthew Astley, Gemma Barson, Gloria Despacio-Reyes, Stephen Fitzgerald, Michael Gray, Ed Griffiths, Deepa Manthravadi, Steve Miller, Christoph Schlaffner, Gosia Trynka, Hendrik Weisser & James Wright
- Program in Computational Biology, Memorial Sloan Kettering Cancer Center, New York, NY, USA Mark Carty, Alvaro Gonzalez, Christina S. Leslie, Yuheng Lu, Alexander R. Perez, Yuri Pritykin & Manu Setty
- 610 Charles E. Young Drive S, Terasaki Life Sciences Building, Room 2000E, Los Angeles, CA, USA Yun-Hua E. Hsiao, Giovanni Quinones-Valdez, Xinshu Xiao & Yi-Wen Yang
- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, Baltimore, MD, USA Michael A. Beer
- Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA Michael A. Beer
- University of California, Santa Cruz, Santa Cruz, CA, USA Joel Armstrong, Alden Deran, Mark Diekhans & Benedict Paten
- Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland Jacqueline Chrast, Anne-Maud Ferreira & Alexandre Reymond
- Centro Nacional de Investigaciones Cardiovasculares (CNIC) and CIBER de Enfermedades Cardiovasculares (CIBERCV), Madrid, Spain Iakes Ezkurdia & Jesus Vazquez
- Spanish National Cancer Research Centre (CNIO), Madrid, Spain Jose Manuel Rodriguez, Michael L. Tress & Alfonso Valencia
- Brunel University London, London, UK Cristina Sisu
- King’s College London, Guy’s Hospital, London, UK Tim J. Hubbard
- ELIXIR Hub, Wellcome Genome Campus, Cambridge, UK Jennifer L. Harrow
- Institute of Cancer Research, London, UK Jyoti S. Choudhary
- Department of Biological Science, Florida State University, Tallahassee, FL, USA Vishnu Dileep, David M. Gilbert, Juan Carlos Rivera-Mulia, Takayo Sasaki & Jared Zimmerman
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota Medical School, Minneapolis, MN, USA Juan Carlos Rivera-Mulia
- Center for Vaccines and Immunology University of Georgia, Athens, GA, USA Michael J. Kulik
- Center for Molecular Medicine and Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, USA Stephen Dalton
- Gift of Life Donor Program, Philadelphia, PA, USA Richard D. Hasz
- American Society for Radiation Oncology, Arlington, VA, USA Judith C. Keen
- National Cancer Institute, National Institutes of Health, Bethesda, MD, USA Helen M. Moore
- Leidos Biomedical, Inc, Frederick, MD, USA Anna Smith
- National Disease Research Interchange (NDRI), Philadelphia, PA, USA Jeffrey A. Thomas
- 4407 Puller Drive, Kensington, MD, USA Peter Good
- Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA, USA Rizi Ai
- Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA Shantao Li
- Michael P. Snyder








